This article is part of a series of practical guides for using the Python data processing library pandas. To see view all the available parts, click here.
To get started with pandas, the first thing you are going to need to understand is how to get data into pandas. For this guide we are going to focus on reading in tabular data (i.e. data stored in a table with rows and columns). If you don’t have some data available but want to try some things out, a great place to get some data to play with is the UCI Machine Learning Repository.
Delimited Files
One of the most common ways you will encounter tabular data, particularly data from an external source or publicly available data, is in the form of a delimited file such as comma separated values (CSV), tab separated values (TSV), or separated by some other character. To import this data so you can start playing with it, pandas gives you the read_csv function with a lot of options to help manage different cases. But let’s start with the very basic case:
import pandas as pd
df = pd.read_csv('path/to/file.csv')
# Show the top 5 rows to make sure it read in correctly
print(df.head())Running this code imports the pandas library (as pd), uses the read_csv function to read in the data and stores it as a pandas DataFrame called df, then prints the top 5 rows using the head method. Note that the path to the file that you want to import can be a path to a file on your computer, or it can be a URL (web address) for a file on the internet. As long as you have internet access (and permission to access the file) it will work like you have the file downloaded and saved already.
When reading the data, unless specified, read_csv will attempt to automatically detect what the delimiting character is (e.g. “,” for CSV). In most cases this works fine, but in cases where it doesn’t, you can use the sep parameter to specify what char to use. For example, if your file is separated with “;” you might do something like:
import pandas as pd
df = pd.read_csv('path/to/file.csv', sep=';')
# Show the top 5 rows to make sure it is correct
print(df.head())OK, what if your file has some other junk above and/or below the actual data like this:
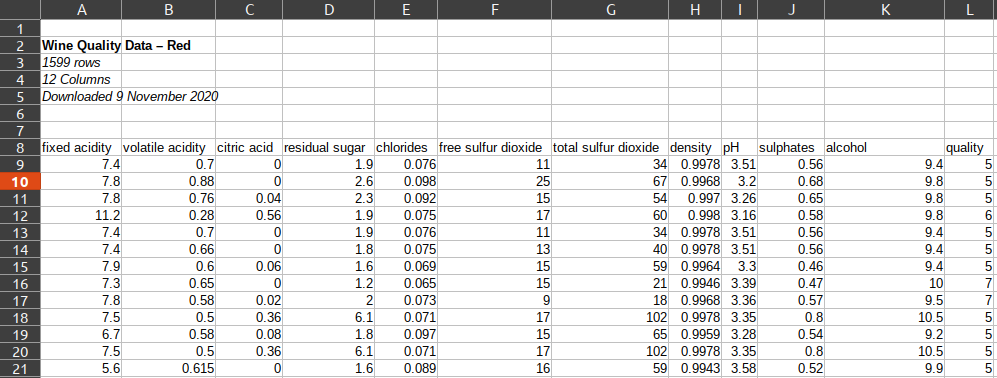
We have two options for working around this, the header parameter and the skiprows parameter:
import pandas as pd
df_1 = pd.read_csv('path/to/file.csv', header=7)
df_2 = pd.read_csv('path/to/file.csv', skiprows=7)
# Both DataFrames produce the same result
print(df_1.head())
print(df_2.head())These are equivalent because setting header=7 tells read_csv to look in row 7 (remember the row numbers are 0 indexed) to find the header row, then assume the data starts from the next row. On the other hand, setting skiprows=7 tells read_csv to ignore the first 7 rows (so rows 0 to 6), then it assumes the header row is the first row after the ignored rows.
Other Useful read_csv Parameters
There are dozens of other parameters to help you read in your data to handle a range of strange cases, but here are a selection of parameters I have found most useful to date:
| Parameter | Description |
|---|---|
skipfooter | Skip rows at the end of the file (note: you will need to set engine=’python’ to use this) |
index_col | Column to set as the index (the values in this column will become the row labels) |
nrows | Number of rows to read in (useful for reading in a sample of rows from a large file) |
usecols | A list of columns to read (can use the column names or the 0 indexed column numbers) |
skip_blank_lines | Skip empty rows instead of reading in as NaN (empty values) |
For the full list of available parameters, checkout out the official documentation. One thing to note is that although there a lot of the parameters available for read_csv, many are focused on helping correctly format and interpret data as it is being read in – for example, interpretation of dates, interpretation of boolean values, and so on. In many/most cases these are things that can be addressed after the data is in a pandas DataFrame, and in some cases, handling these types of formatting and standardization steps explicitly after reading in the data can make it easier to understand for the next person that reads your code.
Excel Data
Pandas also has a nice handy wrapper for reading in Excel data read_excel. Instead of writing your data to a CSV, then reading it in, now you can read directly from the Excel file itself. This function has many of the same parameters as read_csv, with options to skip rows, read in a sample of rows and/or columns, specify a header row and so on.
Databases
If your data is in a tabular/SQL database, like PostgreSQL, MySQL, Bigquery or something similar, your job gets a little bit more complicated to setup, but once that setup is done, it becomes really simple to repeatedly query data (using SQL) from that database directly into a DataFrame where you can do what you want with it.
The first step is to create a connection to the database holding the data. It is beyond the scope of this particular guide, but the library you will almost certainly need to use will be SQLAlchemy or in some cases a library created by the creator of the database (for example, Google has Bigquery API library called google-cloud-bigquery).
Once you have connected to your database, pandas provides three functions for you to extract data into a DataFrame: read_sql_table, read_sql_query and read_sql. The last of these, read_sql, is what’s called a ‘convenience wrapper’ around read_sql_table and read_sql_query – the functionality of both the underlying functions can be accessed from read_sql. But let’s look at the two underlying functions individually to see what the differences are and what options we have.
read_sql_table is a function we can use to extract data from a table in a SQL database. The function requires two parameters table_name – the name of the table you want to get the data from; and con – the location of the database the table is in. With these two parameters, all data from the specified table (i.e. SELECT * FROM table_name) will be returned as a DataFrame:
df = pd.read_sql_table(table_name='table_name', con='postgres:///db_name') read_sql_table does also give you the option to specify a list of columns to be extracted using the columns parameter.
read_sql_query on the other hand allows you to specify the query you want to run against the database.
query = """
SELECT column_1
, column_2
, column_3
FROM table_name
WHERE column_4 > 10
"""
df = pd.read_sql_query(query, 'postgres:///db_name') Obviously writing your own query gives you a lot more flexibility to extract exactly what you need. However, also consider the potential upside in terms of processing efficiency. Doing aggregations and transformations in a database, in almost all cases, will be much faster than doing it in pandas after it is extracted. As a result, some careful query planning can save a lot of time and effort later on.
Other Data Sources
Pandas also has functions for reading in data from a range of other sources, including HTML tables, to SPSS, Stata, SAS and HDF files. We won’t go into them here, but being aware that these options exist is often all you really need to know. If a case arises where you need to read data from these sources, you can always refer to the documentation.
Wrapping Up
We’ve looked at how we can use pandas to read in data from various sources of tabular data, from delimited files and Excel, to databases, to some other more uncommon sources. While these functions often have many parameters available, remember most of them will be unnecessary for any given dataset. These functions are designed to work with the minimum parameters provided (e.g. just a file location) in most cases. Also remember that once you have the data in a DataFrame, you will have a tonne of options to fix and change the data as needed – you don’t need to do everything in one function.
